Schulz Lab
Projects
AI-based precision therapy for heart failure patients
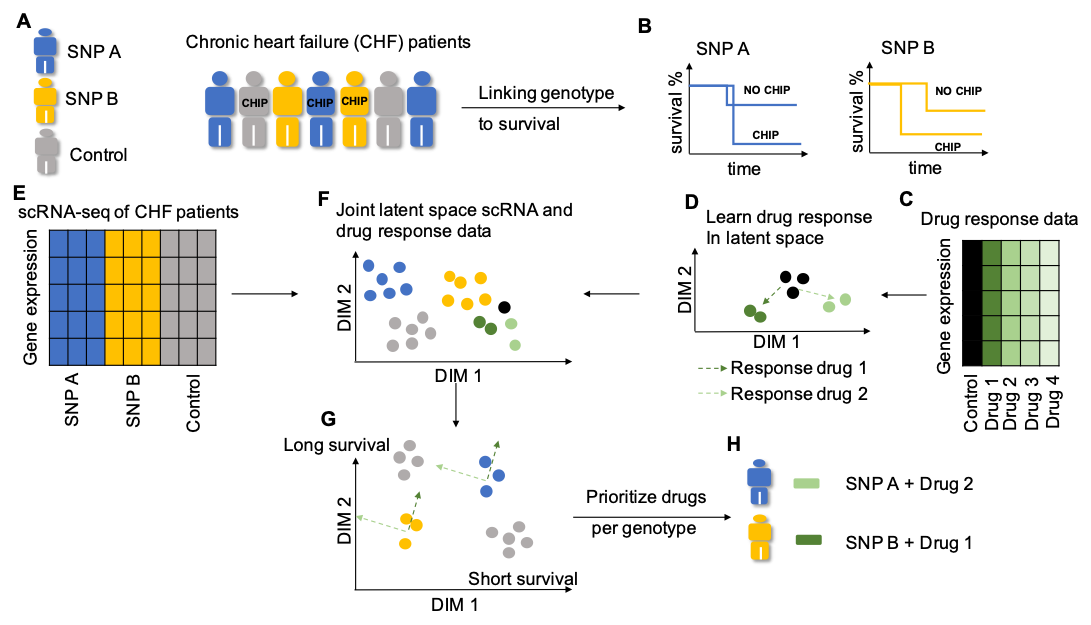
Recent studies show that individual hematopoietic stem/progenitor cells (HSPCs) accumulate somatic mutations as a function of age in healthy individuals. Once a mutation has reached a clone size larger than 2% of the variant allele frequency measured with genomic sequencing, it is called clonal hematopoiesis of indeterminate potential (CHIP). CHIP has been associated to chronic heart failure and atherosclerosis.
Investigating a local cohort of chronic heart failure (CHF) patients, in cooperation with the cardiology department, we have studied the co-occurrence of inherited single nucleotide polymorphisms (SNPs) with the occurrence of CHIP in the genes DNMT3A and TET2. We found a number of inherited SNPs that are associated with a change in survival in the context of CHF patients with CHIP. We are using multi-modal autoencoders to create a joint latent space for the integration of drug response data with single cell RNA-seq data to determine FDA-approved drugs that could provide novel therapies for heart failure treatment. We warmly acknowledge the funding of the computational work on the project by:

Integrative Methods for the Analysis of Gene Regulation
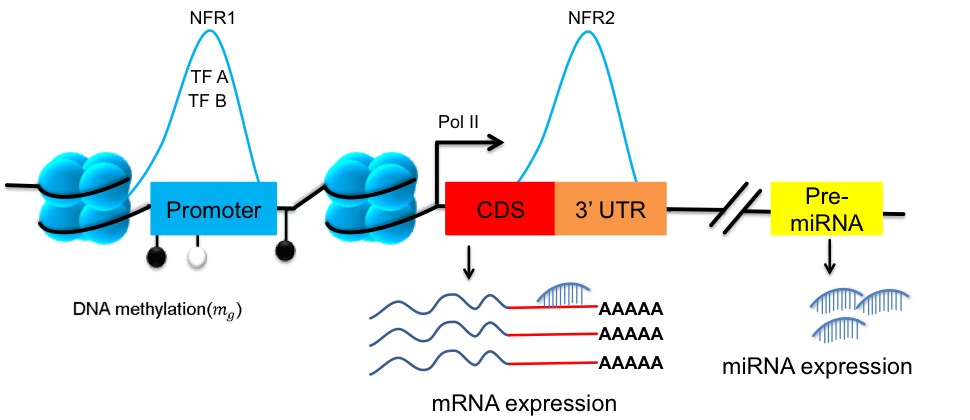
Despite the vast amount of research in that area, the regulation of genes is still not fully understood.
In particular the interplay between regulators that modulate the transcriptional activation of a gene and
post-transcriptional regulators that modulate the abundance of a gene’s product, the mRNA, is neglected in
most system biology studies. With the availability of numerous complete epigenomics datasets our vision is
to produce a comprehensive computational catalogue of gene regulation for each gene in the human genome,
including transcriptional and post-transcriptional regulators, at much higher detail as is currently available.
We greatly acknowledge funding from :


Prioritization of disease relevant transcriptional regulators from single cell data using interpretable machine learning
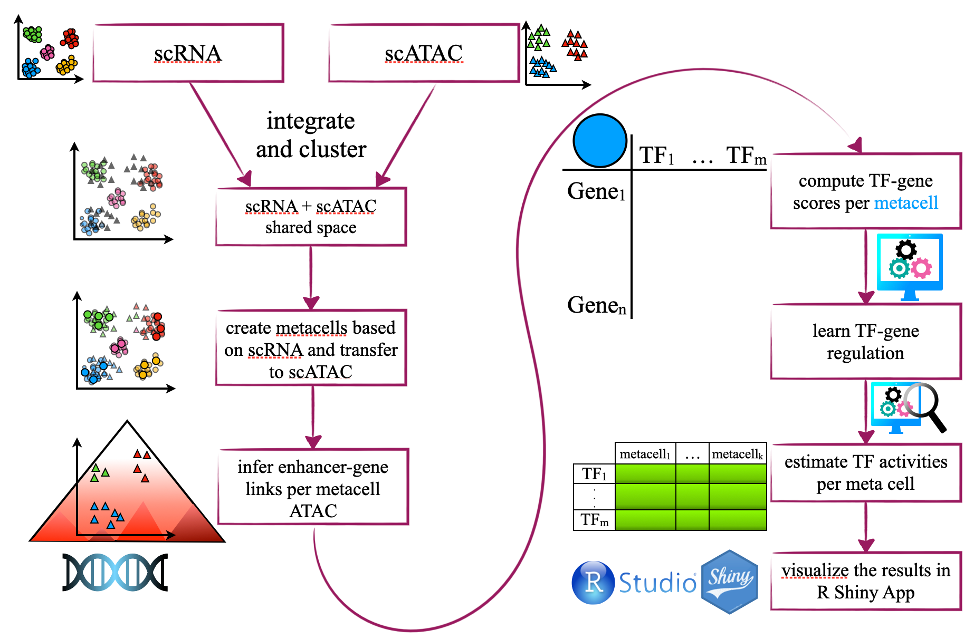
Recent technical developments have enabled the profiling of an individual’s cell transcriptome and epigenome. While single cell measurements provide high resolution and allow the study of gene expression and epigenome changes, they provide noisy and sparse measurements of a cell’s activity, in particular for the commonly used 10X sequencing platform. We have been developing machine learning methods for the analysis of single cell RNA and epigenome data that allow prioritization of transcriptional regulators of interest. We have developed a statistical framework with an R shiny-based application, called GAZE, that guarantees a comprehensive analysis of single-cell data in an integrative fashion. Using dimensionality reduction and unsupervised machine learning, sparsity of single cell data is reduced by aggregation to so-called metacells. Then supervised sparse regression is used to learn associations between TF binding predictions in enhancer/promoters and gene expression in a per-gene fashion extending our tree-guided multitasking method to associate transcription factor regulation of genes using single cell data.
Reconstruction of dynamic regulatory networks
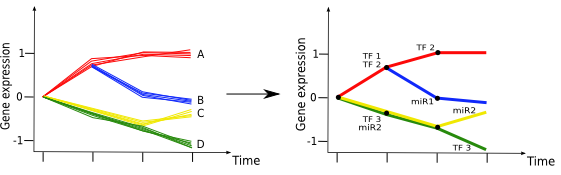
Current models of gene regulatory networks are often constructed as a static snapshot of the regulatory wiring in cells. We are working on methods that can dynamically rewire the network connections modeling transcriptional and posttranscriptional factors through the integration of binding data (e.g. Chip-Seq) and gene expression data. In addition, we are enhancing these methods to utilize transcript expression level measurements with RNA-Seq to improve the resolution for reconstruction of dynamic regulatory networks.
D Gérard, F Schmidt, A Ginolhac, M Schmitz, R Halder, P Ebert, MH Schulz, T Sauter, L Sinkkonen
Temporal epigenomic profiling identifies AHR and GLIS1 as super-enhancer controlled regulators of mesenchymal multipotency,
Nucleic Acids Research 2019 [full text]
MH Schulz, KV Pandit, CL Lino Cardenas, N Ambalavanan, N Kaminski and Z Bar-Joseph
Reconstructing dynamic microRNA-regulated interaction networks
PNAS 2013 [full text]
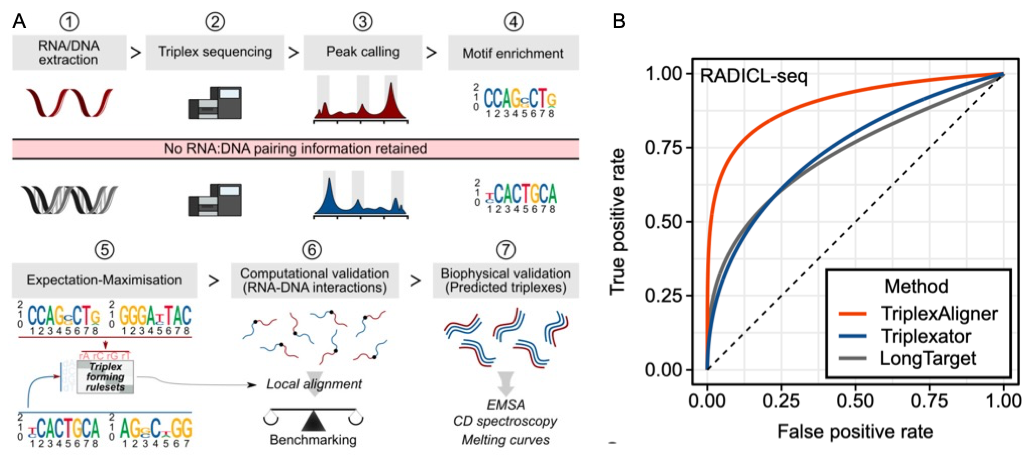
An unbiased approach reveals a universal code for RNA·DNA:DNA interaction
RNA·DNA:DNA triple helix (triplex) formation is a form of RNA-DNA interaction, which regulates gene expression, but is difficult to study experimentally in vivo. This makes accurate computational prediction of such interactions highly important in the field of RNA research. Current predictive methods use canonical Hoogsteen base pairing rules, which whilst biophysically valid, may not reflect the plastic nature of cell biology. We have developed the first unbiased optimization approach to learn a probabilistic model describing RNA-DNA interactions directly from motifs derived from triplex sequencing data. We find that there are several stable interaction codes, including Hoogsteen base pairing and novel RNA-DNA base pairings, which agree with in vitro measurements of triplex binding. We implemented these findings in TriplexAligner, a program that uses the determined interaction codes to predict triplex binding. TriplexAligner predicts RNA-DNA interactions identified in all-to-all sequencing (RADICL-seq) data more accurately than all previously published tools, and also predicts previously studied triplex interactions with known regulatory functions. Our work is an important step towards better understanding of triplex formation and allows genome-wide analyses of RNA-DNA interactions.
We acknowledge funding by 
T Warwick, et al.
A universal model of RNA.DNA:DNA triplex formation accurately predicts genome-wide RNA-DNA interactions,
Briefings in Bioinformatics, 2022 [full text]